Man and Machine
It's a tired joke I've been using for decades now: I've written code to cure cancer! Actually, it causes cancer, but with a little more debugging, I'm sure I can reverse that. The reality, of course, is that we can make machines do so much, but so much of that is really insignificant. Sure, programming mission-critical business systems requires solving some interesting problems, but those problems often pale in comparison with many others in society. So perhaps every programmer's dream is to write that code that actually will help cure diseases or solve another similar ill.
Protein Folding
Protein folding research is one such area where some new, creative software solutions are being applied to significant, hard problems. There's a lot we know about protein assembly (for example, diseases arise from incorrect folding), and a lot we don't know, like how proteins fold so well and how they sometimes misfold. Answers to these questions might lead to cures for cystic fibrosis, HIV, Alzheimer's disease, Parkinson's disease, Huntington's disease, Mad Cow disease, congenital emphysema, allergies, and many cancers. Yet simulating folds to collect research data is computationally expensive: it can take around 30 CPU years to simulate one folding sequence.
So what does one do when hitting a computational wall? Either get more CPUs (quantity) or better ones (quality).
More Brains
Projects such as Stanford University's Folding@home (and others like it) are taking the more CPUs approach. They use distributed computing (not unlike Berkeley's SETI@home) to farm out simulation work to nearly a half million volunteer PCs around the world.
Better Brains
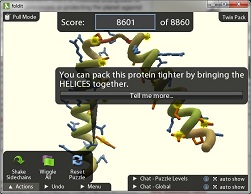
The University of Washington's (UW) Foldit project takes the better CPUs approach: specifically, the human mind. Many computer gamers possess strong, intuitive 3-D problem solving skills: skills that can be used to solve complex protein folding puzzles faster than computers can. The challenge is finding and motivating these protein-folding savants, something UW accomplishes by making a game of it, and a world-wide competition.
Foldit is a 3-D protein folding puzzle-solving game. Think of it as Tetris for protein designers. Getting started with it is quick and easy; one click downloads and installs from the web site. The Intro puzzles serve as a tutorial: just start solving puzzles, letting the hints guide the way. The program does an excellent job of directing you toward proper folds, with 3-D rotation, bubble hints, red stars marking problems, and a sliding score that updates as you move. It's a study in transitional interfaces; for example, as you progress, it unfolds (pun intended) new tools you can use.
I suppose Foldit's one weakness is its strength: you end up making structural changes intuitively, without considering the science behind them. "All this science I don't understand," indeed, but that's okay: the top five folders in a recent study had no more than a high-school level knowledge of biochemistry. To quote one of the designing professors, "Some people are just able to look at the game and in less than two minutes, get to the top score. They can't even explain what they're doing, but somehow they're able to do it."
So if you have a knack for quickly solving 3-D puzzles, give Foldit a try. Who knows, you just might cure cancer.